Translate this page into:
Biochemical and molecular characterization of proteolytic bacterial strains isolated from Jazan region, KSA with the application as an antibacterial agent

*Corresponding author: Bander Mohammed Al-Thobaiti, Department of Biology, Jazan University, Jazan, Kingdom of Saudi Arabia. bander_m_j@hotmail.com
-
Received: ,
Accepted: ,
How to cite this article: Al-Thobaiti BM, Abada E, El-Gayar K. Biochemical and molecular characterization of proteolytic bacterial strains isolated from Jazan region, KSA with the application as an antibacterial agent. Mod J Microb Biol 2021;3(1):1-17.
Abstract
Objectives:
Biochemical and molecular characterization of proteolytic bacterial strains isolated from Jazan region, KSA with the application as an antibacterial agent.
Materials and Methods:
Three samples were collected from extreme environment, Jazan, KSA. Skim milk nutrient agar medium was used for protease screening for several colonies by streaking method at 37°C. API biochemical kit was used to characterize the three isolates using some selective media. The genetic identification was done using 16S rRNA gene sequencing. The sensitivity of the tested strains;Staphylococcus aureus, Escherichia coli, and Klebsiella pneumoniae against the extracellular crude protease enzyme produced from the three isolated bacteria and different antibiotics was done.
Results:
The isolates were identified as Bacillus subtilis, Bacillus licheniformis, and Bacillus cereus. B. cereus and B. licheniformis recorded high sensitivity (71%) against most antibiotics, in addition, B. subtilis showed resistance to Aztreonam only. It was found a considerable increase in the level of both of protease activity (units/ml) and bacterial growth (colony-forming units/ml) of the cultures that were directed by the B. subtilis and B. licheniformis up to 37°C then decreased at 45°C. On the contrary, the growth of B. cereus and its activity gradually increased up to 45°C. The enzyme activity and bacterial growth of B. subtilis and B. cereus strains were increased at alkaline medium. However, B. licheniformis gave the highest growth and activity at neutral pH. In addition, it was found that the enzyme activity and bacterial growth of B. subtilis were reached to the maximum at 5% NaCl. However, the maximum bacterial growth and enzyme activity for B. licheniformis and B. cereus was at 2% NaCl. It was found high effect on inhibiting the growth of pathogenic bacteria using 5 μl of crude enzyme with specific enzyme activity 73, 76, and 92 (units/ml)/(mg protein/ml) for B. subtilis, B. licheniformis, and B. cereus, respectively. All pathogenic bacteria were totally inhibited with 10 μl of crude enzyme.
Conclusion:
The potential Bacillus proteases can promote new industry as antimicrobial agents.
Keywords
Proteolytic Bacillus strains
Pathogenic bacteria
Antibacterial agent
INTRODUCTION
Proteolysis considers one of the most important biological reactions. These enzymes are of wide distribution, and they perform significant biological processes.[1] Proteases are produced in animals, plants, bacteria, archaea, and viruses. Among the different sources of proteases, bacterial protease was the most significant compared with animal, fungi, and plant protease. Bacillus species were specific producers of extracellular protease.[2]
Bacterial diseases become a serious problem in public health. Antibiotic resistance as become one of the largest worldwide anxieties. Resistance to antibiotics occurs because of the changing nature of bacteria that no longer can be killed. Efficacy of the drug lost. Bacterial strains that are resistant to different antibiotics will multiply then spread so that they become more serious.[3] Recent reports indicated that various microbial enzymes might be used as potential matrix-degrading therapeutic agents.[4]
Proteases are involved in many aspects of human biology. For example, in the small intestine, proteases digest dietary proteins to allow absorption of amino acids. Other processes mediated by proteases include blood coagulation, immune function, maturation of prohormones, bone formation, programmed cell death, and the recycling of cellular proteins that are no longer needed. Proteases also offer a valuable target in many therapeutic settings, including cancer, Alzheimer’s, and viral infection. A matrix metallopeptidase-9 plays a role in angiogenesis and is a therapeutic target for cancer. Because of their significance in the pathology of disease, proteases are a relevant drug target class. Proteases activity is central to diverse physiological cascades throughout biology. Some are essential for coagulation, while others contribute to cancer pathology.[5,6]
Bacterial diseases become a serious problem in public health. Antibiotic resistance becomes one of the largest worldwide anxieties. Resistance to antibiotics occurs because of the changing nature of bacteria that no longer can be killed. Efficacy of the drug lost. Bacteria that are resistant to antibiotics will multiply and spread so that they become more dangerous.[3] Recent reports indicated that various microbial enzymes might be used as potential matrix-degrading therapeutic agents.[4]
MATERIALS AND METHODS
Samples collection
Three samples were collected from different extreme environment [Figure 1], Jazan, Kingdom of Saudi Arabia. Samples were assembled in 500 mL glass screw cap bottles and brought to the laboratory. These sources of the bacterial isolation were, Salty soil from Farasan island (S), Al-Harth Hot spring soil (H), and Salty water, Red Sea, Farasan Island (W).

- Sources locations for the isolated samples.
Isolation, purification, and screening of proteolytic bacteria
One milliliter of the bacterial sample was taken and diluted with pre-sterilized physiological saline solution (under aseptic conditions) to reach to appropriate dilutions. A 100 μl of each dilution was plated on milk agar (M.A.) in triplicates. The plates were incubated O.N. at 37°C and the number of developed colonies was processed. After incubation, the total bacterial count was recorded as colony-forming units (CFU) mL−1 as described earlier. The developed colonies were repeatedly streaked on NA plates to isolate of pure cultures.[7]
Skim milk nutrient agar medium was used for protease screening for several colonies by streaking method using sterile toothpick and incubated at 37°C. Colonies forming transparent zones, because of partial hydrolysis of milk casein, were selected.[8]
The gelatin hydrolysis test was used to detect the ability of isolated microorganism to produce the enzyme gelatinase. The procedures were done by inoculation a heavy inoculum by stabbing on the tube containing nutrient gelatin medium. After that, incubation the inoculated tube along with an uninoculated medium which utilized as a control tube at 37°C for 14 days. Removing the tubes daily from the incubator and placing in refrigerator (4°C) for 15–30 min until control is gelled. Every day the same method repeats to check for gelatin liquefaction.[9]
Characterization of bacterial isolates
Gram staining was done using a commercially kit (Quimica Clinica Aplicada, S.A., Amposta, Spain) according to the standard method.[10]
Catalase mediates the breakdown of hydrogen peroxide H2O2 into oxygen and water. To find out if a particular bacterial isolate is able to produce catalase enzyme, a small inoculum of a bacterial isolate is mixed into hydrogen peroxide solution (3%) and is observed for the rapid elaboration of oxygen bubbles.[11]
Biochemical characterization of bacterial isolates
Biochemical characterization of the proteolytic bacterial isolates was performed using commercially available identification systems API, BioMérieux.[12]
Antibiotics susceptibility test
A single colony of the isolates was picked using a sterile loop and streaked on the Muller-Hinton agar plate. A filter paper disk contained the antibiotics; Cefoxitin (FOX, 30 μg), Aztreonam (ATM, 30 μg), Cefotaxime (CTX, 30 μg), Ceftazidime (CAZ, 30 μg), Amoxicillin/Clavulanate (AUG, 30 μg), Meropenem (MEM, 10 μg), Amikacin (AK, 30 μg), Cefepime (CPM, 30 μg), Piperacillin/Tazobactam (PTZ, 110 μg), Imipenem (IMI, 10 μg), Tetracycline (T, 30 μg), Teicoplanin (TEC, 30 μg), Neomycin (NE, 30 μg), Mupirocin (MUP, 200 μg), Mupirocin (MUP, 5 μg), Rifampicin (RP, 5 μg), Erythromycin (E, 15 μg), Vancomycin (VA, 30 μg), Clindamycin (CD, 2 μg), Trimethoprim/Sulfamethoxaz (TS, 25 μg), Cefuroxime (CXM, 30 μg), Trimethoprim (TM, 5 μg), Ciprofloxacin (CIP, 5 μg), Amoxicillin (AX, 10 μg), Cefazolin (CZ, 30 μg), Penicillin (PG, 10 μg), Fucidin Acid (FC, 10 μg), and Nitrofurantoin (NI, 300 μg) separately were dispensed onto the plate. Using sterilized forceps, each disk was placed on the agar medium to ensure that the disk is attached and fixed on the agar. Muller-Hinton agar medium containing antibiotic disks was incubated O.N. at 35°C in incubator.[13]
Identification using 16S rRNA
The selected proteolytic bacterial isolates were identified using 16S rRNA gene sequencing technique. The genomic DNA was extracted using TIANamp genomic DNA kit (Tiagen, Korea), according to its manufacturer instructions. The primers used in amplification of the 16S rRNA gene were forward primer (F; AGA GTT TGA TCC TGG CTC AG) and reverse primer (R; GGT TAC CTT GTT ACG ACT T).[14] PCR was performed according to Sambrook and Russel, 2001,[15] with some modifications for 35 cycles under the following conditions: Denaturation step at 94°C for 40 s, annealing step at 55°C for 1 min, extension step at 72°C for 2 min, and final extension at 72°C for 10 min. Sequencing of the amplified fragments was done at Macrogen biotechnology company. Cycle Sequence PCR fragments were purified by cycle sequencing purification, using big dye X terminator (applied Bio system). Sequence analysis was performed using the sequence alignment software BLASTn with the national center for biotechnology information (NCBI) database, http://www.ncbi.nlm.nih.gov/.
Effect of some factors on the bacterial growth and protease activity
Bacterial cells were activated by growing them O/N on M.A. plate at 37°C. Ten milliliter of Casein soyabean broth medium were inoculated with several recently growing colonies (fresh colonies) after which cells were allowed to grow for 24 h at 30°C with shaking at 150 rpm. To study the effect of temperature on the bacterial growth and protease activity, three flasks containing 100 ml of fresh Casein soyabean broth medium were inoculated with 100 μl of the above culture, for each. The new culture was allowed to grow at 30, 37, and 45°C with shaking for 24 h. Activated bacterial cells were used to inoculate Casein soyabean broth medium with different pH (4, 7, and 9). Cultures were grown at 30°C with shaking at 150 RPM for 24 h. In another experiment to study the effect of salinity on the bacterial growth and protease activity, Casein soyabean broth media were supplemented with 2, 5, and 10% Na Cl. In addition, all cultures were grown at 30°C with shaking at 150 RPM for 24 h. For all cultures, at the indicated time, 2 ml of the growing culture was taken and measured the O.D. at Ab. 600 nm. To get cells free supernatant, 1.5 ml of the culture was centrifuged in a microcentrifuge at 8000 rpm for 5 min. The supernatants were then used as crude enzyme to determine the protease activity.[16]
Determination of the protease activity
The activity of the protease was determined according to the method of Sutar et al. 1986.[17] Reaction mixtures (2 ml) contained 10 mg casein, 200 μmol sodium carbonate buffer, pH 9.7 and 0.1 ml of the supernatants. Reactions were carried out at for 30 min then were terminated by the addition of 2.6 ml 5% (w/v) trichloroacetic acid (TCA), and 0.4 ml 3.3 M HCl. Reactions were then kept on ice for at least 1 h after which they were centrifuged for 30 min at 4000 rpm. The absorbance of the TCA soluble fractions was measured at 280 nm. One unit of enzyme activity was equal to the amount of enzyme that liberates 1 μmol of tyrosine from casein/30 min at 37°C. A standard curve using the amino acid tyrosine was established.
Determination of soluble protein content
The content of soluble proteins was estimated using the bicinchoninic acid (BCA) assay kit (Sigma-Aldrich BCA-1, B9643), with bovine serum albumin as a standard.[18]
Antimicrobial activity test
The sensitivity of three different pathogenic bacteria against the extracellular crude protease enzyme produced from the three isolated bacteria was done, according to the previous studies.[19,20] The three pathogens strains were isolated and identified previously, Biology Department, Faculty of Science, Jazan University.[21] These pathogenic isolates were one Gram-positive bacterial strain, Staphylococcus aureus. In addition, Gram-negative bacterial strains, Escherichia coli, and Klebsiella pneumoniae.
The isolated bacterial cells were activated by growing them overnight on Nutrient agar supplemented with skim milk at 37°C. Ten milliliter of Casein Soyabean Digest Broth were inoculated with several recently growing colonies (fresh colonies) after which cells were allowed to grow for 3 h at 37°C with shaking at 150 rpm. A 100 ml of fresh Casein Soyabean Digest Broth were inoculated with 1 ml of the above culture. The new culture was allowed to grow overnight at 37°C with shaking. At the indicated time, CFU/ml was determined. In addition, 1.5 ml of the growing culture was taken and centrifuged in a microcentrifuge at 7000 rpm for 2 min. The supernatants were then used as crude enzyme to determine the activity of protease, protein content as well as to test as antimicrobial agents. Ten microliters of activated pathogen cells were inoculated to 10 ml of Casein Soyabean Digest Broth, supplemented with 5 or 10 μl crude enzyme extract. All cultures were incubated O/N at 37°C. At the indicated time, CFU/ml and measurement of turbidity at 600 nm were determined to detect the antimicrobial activity of the tested proteolytic isolated strains.
RESULTS
Isolation of proteolytic bacterial strains
Three samples were collected from different extreme environment, Jazan, Kingdom of Saudi Arabia as mentioned before. Isolation of bacterial colonies was done on M.A. with incubation at 30°C [Figure 2a]. The total bacterial count for salty soil, Hot spring, and Sea water samples was 1.2 × 107, 6 × 103, and 2 × 105 CFU/ml, respectively. Among the different isolates, three isolates (one bacterial isolate from each resource) with the maximum zone of clearance on skim M.A. and high capacity for the liquefaction of gelatin were selected [Figures 2b and c]. Bacterial isolates were purified using streaking technique on skim M.A. The three strains were coded as follow: S: Strain isolated from Salty soil, H: Strain isolated from Al-Harth Hot spring, and W: Strain isolated from Seawater.
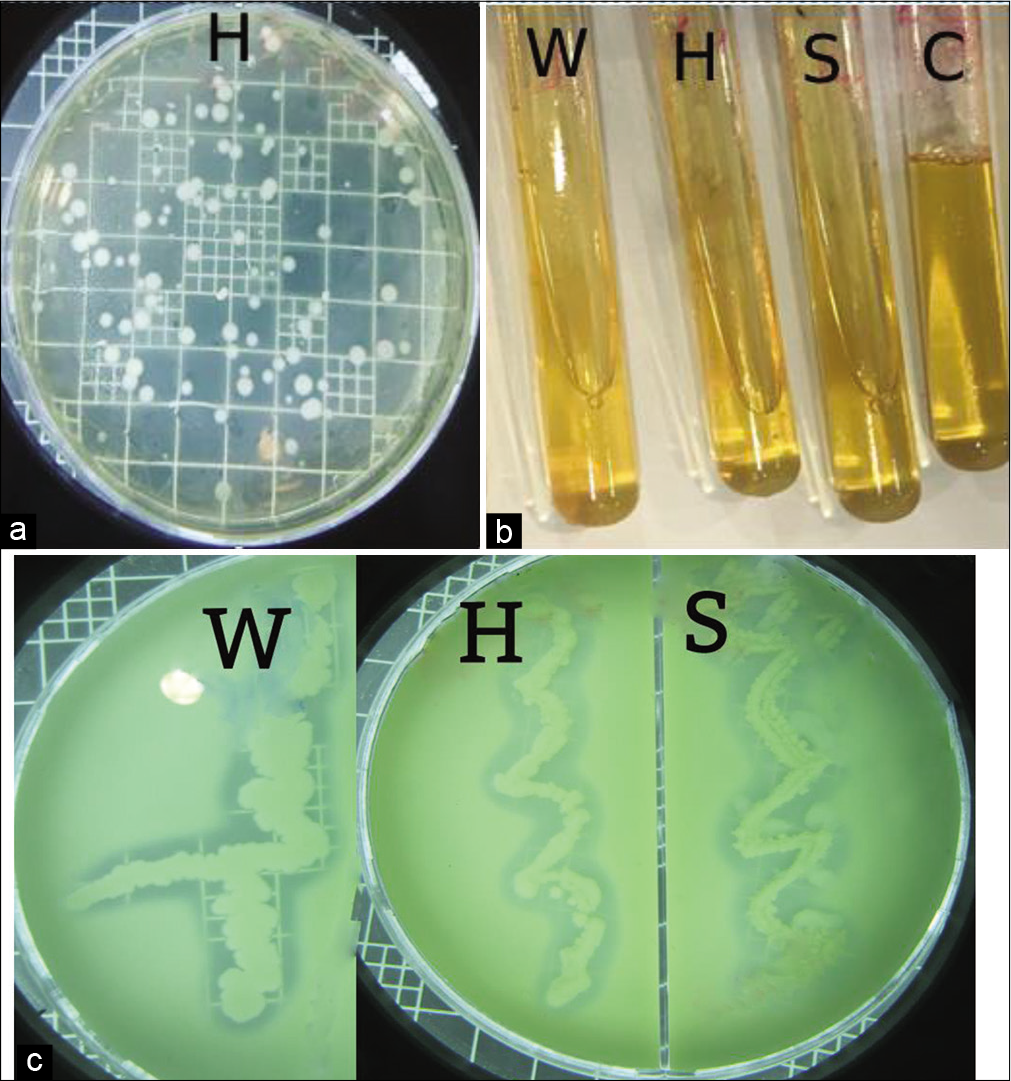
- (a) Isolation on milk agar with incubation O/N at 30°C (Hot spring sample), (b) Protease qualitative determination of the bacterial isolates by gelatin liquefaction, where C: control (gelatin without inoculum), S: Isolated from Salty soil, H: Isolated from Hot spring and W: Isolated from Seawater, (c) Protease qualitative determination of the bacterial isolates on skim milk nutrient agar, Where, S: Isolated from Salty soil, H: Isolated from Hot spring and W: Isolated from Seawater.
Characterization of bacterial strains
After purification of selected maximum proteolytic strains, they were observed using the light microscope. The morphology graph of isolated bacteria showed that all isolates were Gram-positive and rod-shaped bacteria.
Catalase test was performed for all isolates. All of them were catalase-positive bacteria. Therefore, they were strict aerobes as well as facultative anaerobes.
Using API biochemical kit, the three of the Gram-positive rods isolated from extreme environment, Jazan, KSA, were characterized using some selective media [Table 1].
| No. | Test | Reaction | Isolate S | Isolate H | Isolate W |
|---|---|---|---|---|---|
| 1 | ONPG | β-galactosidase | − | − | − |
| 2 | ADH | Arginine dihydrolase | + | + | + |
| 3 | LDC | Lysine decarboxylase | + | + | − |
| 4 | ODC | Ornithine decarboxylase | + | + | − |
| 5 | CIT | Citrate utilization | − | − | − |
| 6 | H2S | H2S production | − | − | − |
| 7 | URE | Urea hydrolysis | + | − | − |
| 8 | TDA | Tryptophan deamination | + | + | + |
| 9 | IND | Indole production | + | + | − |
| 10 | VP | Acetoin production | + | + | + |
| 11 | GEL | Gelatin hydrolysis | + | + | + |
| 12 | GLU | Glucose fermentation | − | − | − |
| 13 | MAN | Mannitol | − | − | − |
| 14 | INO | Inositol | − | − | − |
| 15 | SOR | Sorbitol | − | − | − |
| 16 | RHA | Rhamnose | + | + | + |
| 17 | SAC | Sucrose | − | − | − |
| 18 | MEL | Melibiose | + | + | + |
| 19 | AMY | Amygdalin | − | − | − |
| 20 | ARA | Arabinose fermentation | + | + | − |
| 21 | ESC | Aesculin hydrolysis | + | + | + |
| 22 | PYRA | Pyrrolidonylaryl-amidase | − | + | − |
| 23 | αGAL | α-galactosidase | − | + | − |
| 24 | βGUR | β-glucuronidase | − | + | − |
| 25 | βGAL | β-galactosidase | − | + | − |
| 26 | PAL | Alkaline phosphatase | − | + | − |
| 27 | LAP | Leucine arylamidase | − | + | − |
| 28 | RIB | Ribose fermentation | + | + | + |
| 29 | MAN | Mannitol fermentation | − | − | − |
| 30 | SOR | Sorbitol fermentation | − | − | − |
| 31 | LAC | Lactose fermentation | − | + | − |
| 32 | TRE | Trehalose fermentation | − | − | + |
| 33 | INU | Inulin fermentation | + | − | − |
| 34 | RAF | Raffinose fermentation | − | − | − |
| 35 | AMD | Starch fermentation | + | − | + |
| 36 | GLYG | Glycogen fermentation | + | − | + |
+: Positive, −: Negative
Antibiotic susceptibility test
Susceptibility test is used when microbe is unable to grow in the presence of one or more antimicrobial drugs. Testing is used to determine the potential effectiveness of specific antibiotics on the bacteria and/or to determine if the bacteria have developed resistance to certain antibiotics. The obtained results [Table 2] show the antibiotic susceptibility pattern of the bacterial isolates in this study. The bacterial isolate (W) recorded high sensitivity (71%) with different degrees against most antibiotics, but also showed resistance to the following antibiotics: PG, AX, ATM, TM, CXM, MUP, CPM, and CAZ. Furthermore, the bacterial strain (S) showed resistance to ATM only, and it was highly sensitive to the rest of the antibiotics (96%). In the meantime, the bacterial strain (H) showed sensitivity against most antibiotics (71%) and showed resistance to the following antibiotics: ATM, T, TEC, MUP 5, CD, AX, PG, and FC.
| No. | Antibiotic | Conc.(g/disc) | Isolate W | Isolate S | Isolate H | |||
|---|---|---|---|---|---|---|---|---|
| Status | Halo-zone mm | Status | Halo-zone mm | Status | Halo-zone mm | |||
| 1 | Cefoxitin | 30 | S | 12 | S | 31 | S | 26 |
| 2 | Aztreonam | 30 | R | 0 | R | 0 | R | 0 |
| 3 | Cefotaxime | 30 | S | 9 | S | 23 | S | 30 |
| 4 | Ceftazidime | 30 | R | 0 | S | 18 | S | 20 |
| 5 | Amoxicillin/Clavulanate | 30 | S | 10 | S | 30 | S | 14 |
| 6 | Meropenem | 10 | S | 28 | S | 40 | S | 36 |
| 7 | Amikacin | 30 | S | 30 | S | 30 | S | 20 |
| 8 | Cefepime | 30 | R | 0 | S | 30 | S | 33 |
| 9 | Piperacillin/Tazobactam | 110 | S | 27 | S | 30 | S | 27 |
| 10 | Imipenem | 10 | S | 37 | S | 46 | S | 39 |
| 11 | Tetracycline | 30 | S | 18 | S | 22 | R | 0 |
| 12 | Teicoplanin | 30 | S | 18 | S | 18 | R | 0 |
| 13 | Neomycin | 30 | S | 25 | S | 25 | S | 17 |
| 14 | Mupirocin | 200 | S | 10 | S | 39 | S | 25 |
| 15 | Mupirocin | 5 | R | 0 | S | 30 | R | 0 |
| 16 | Rifampicin | 5 | S | 14 | S | 20 | S | 13 |
| 17 | Erythromycin | 15 | S | 29 | S | 35 | S | 13 |
| 18 | Vancomycin | 30 | S | 18 | S | 25 | S | 9 |
| 19 | Clindamycin | 2 | S | 22 | S | 20 | R | 0 |
| 20 | Trimethoprim/Sulfamethoxaz | 25 | S | 18 | S | 35 | S | 22 |
| 21 | Cefuroxime | 30 | R | 0 | S | 21 | S | 25 |
| 22 | Trimethoprim | 5 | R | 0 | S | 35 | S | 23 |
| 23 | Ciprofloxacin | 5 | S | 22 | S | 35 | S | 31 |
| 24 | Amoxicillin | 10 | R | 0 | S | 31 | R | 0 |
| 25 | Cefazolin | 30 | S | 8 | S | 37 | S | 13 |
| 26 | Penicillin | 10 | R | 0 | S | 40 | R | 0 |
| 27 | Fucidin acid | 10 | S | 13 | S | 28 | R | 0 |
| 28 | Nitrofurantoin | 300 | S | 22 | S | 25 | S | 25 |
R: Resistance, S: Sensitive
16S rRNA identification
Three Gram-positive Bacilli bacteria with maximum zone of clearance on skim milk, and high capacity for the liquefaction of gelatin due to proteolytic activity were selected to identify. After DNA extraction, followed with PCR, the molecular identification of the bacterial isolates was performed using 16S rRNA gene sequencing. The obtained DNA sequences were aligned at the NCBI database. [Table 3] shows the accession no, identity percentage and identification of the isolates. The sequences and phylogenetic tree of the selected strains identified 16S rRNA is shown at [Figures 3-5]. The Bacilli species were identified as Bacillus subtilis (isolated from salty soil.), Bacillus licheniformis (isolated from Al-Harth hot spring), and Bacillus cereus (isolated from seawater).

- The homology of partial DNA sequences of the 16S rRNA gene of the bacterial strain isolated from Salty water, Jazan, KSA (code S) and the corresponding gene of Bacillus subtilis strain (accession number KU229984).

- Phylogenetic tree of the 16S rRNA sequence results of Bacillus subtilis.

- The homology of partial DNA sequences of the 16S rRNA gene of the bacterial strain isolated from Al-Harth hot spring, Jazan, KSA (code H) and the corresponding gene of Bacillus licheniformis strain (accession number MK622385).
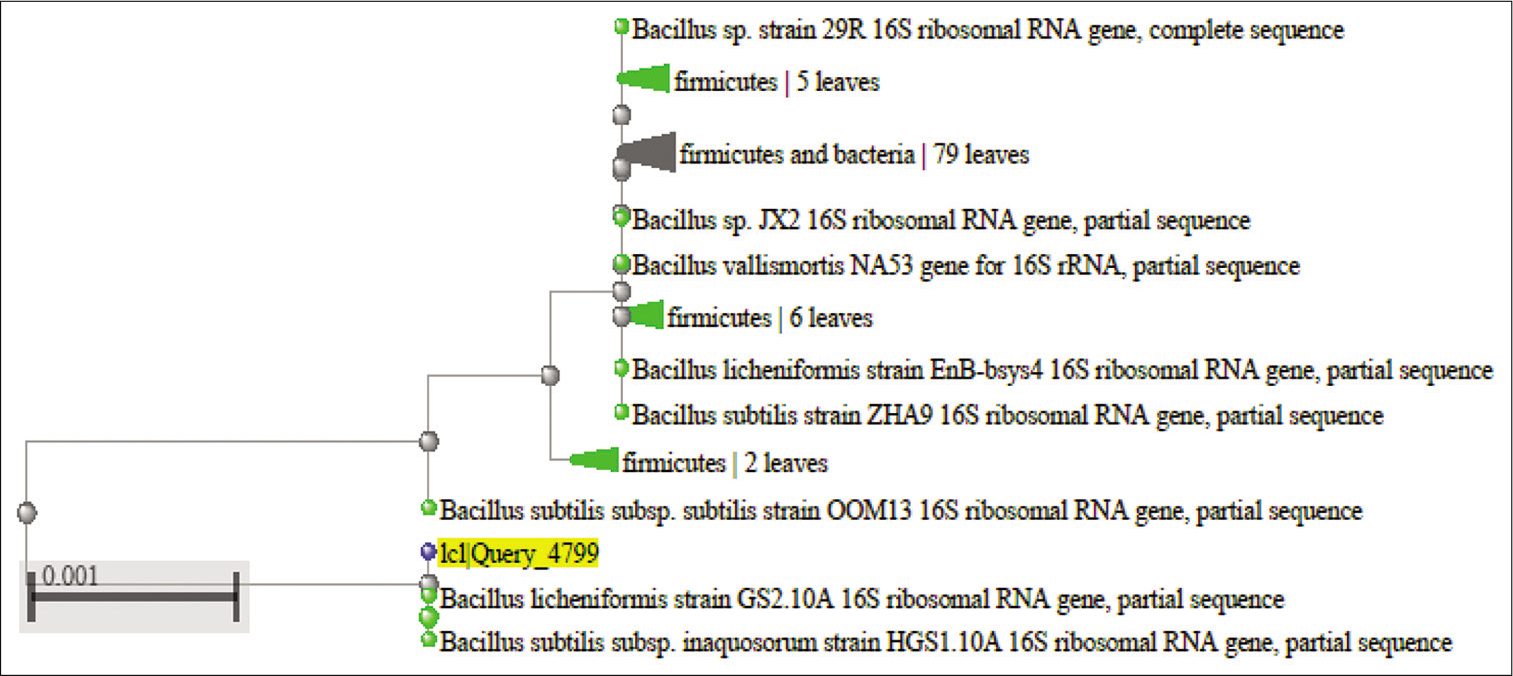
- Phylogenetic tree of the 16 S rRNA sequence results of Bacillus licheniformis.
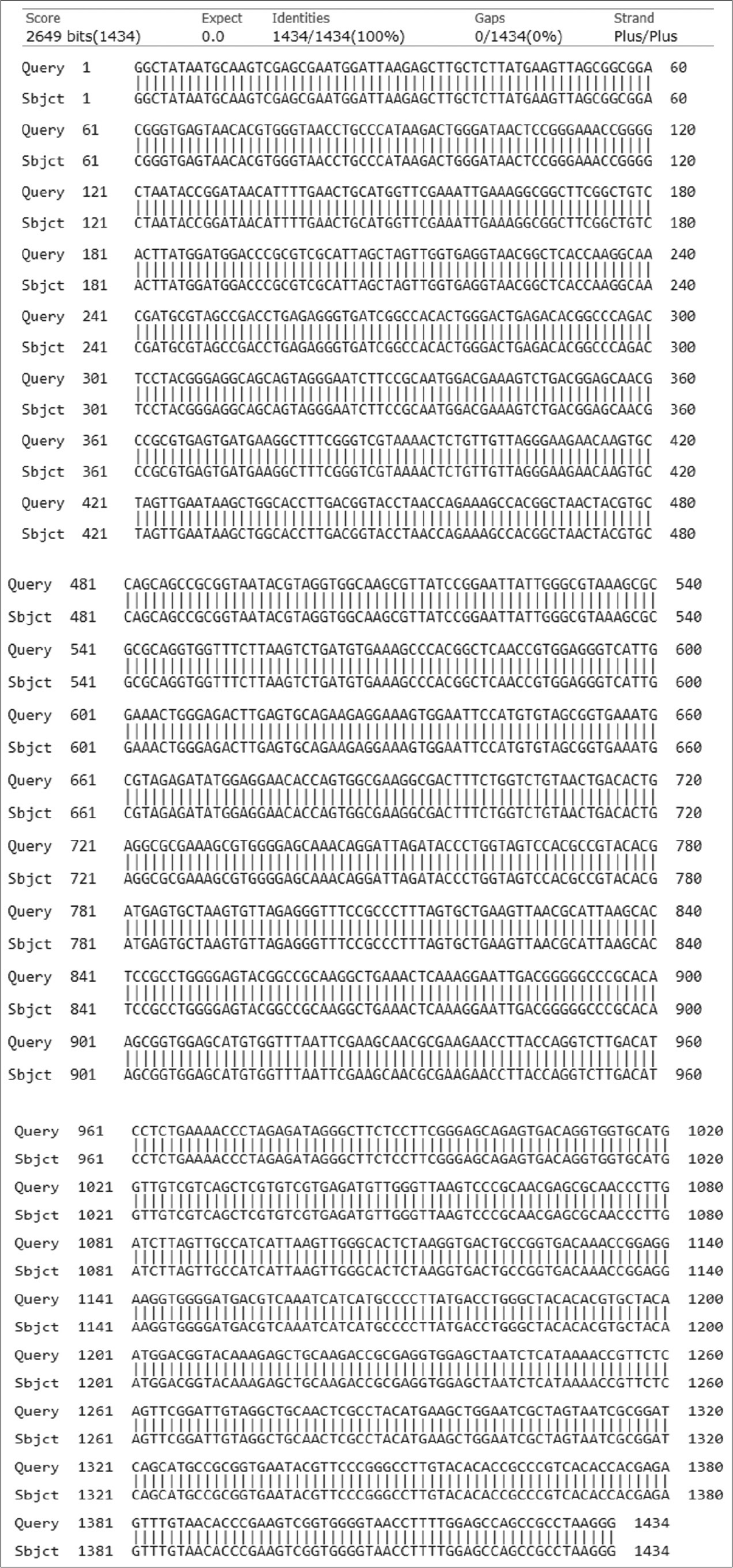
- The homology of partial DNA sequences of the 16S rRNA gene of the bacterial strain isolated from Farasan Island sea water, Jazan, KSA (code W) and the corresponding gene of Bacillus cereus strain (accession number KJ534410).

- Phylogenetic tree of the 16 S rRNA sequence results of Bacillus cereus.
| Isolation resource | Sequence length | Accession number | Identity (%) | Identification |
|---|---|---|---|---|
| Isolated S | 1483 bp | KU229984 | 96.75 | Bacillus subtilis |
| Isolated H | 1095 bp | MK622385 | 93.49 | Bacillus licheniformis |
| Isolated W | 1434 bp | KJ534410 | 96.43 | Bacillus cereus |
Factors affecting on bacterial growth and protease production
Some factors affecting on bacterial growth and protease production were studied as mentioned above. The bacterial growth was monitored by calculating (CFU/ml). The protease activity was monitored using casein method as mentioned before regarding to tyrosine standard curve. One unit of enzyme activity can defined as the amount of enzyme that liberates 1 μmol tyrosine/30 min at 37°C using casein as a substrate.
[Figures 6a and b] illustrate that there is a considerable increase in the level of both of protease activity (Units/ml) and bacterial growth (CFU/ml) of the culture that was directed by B. subtilis at 37°C. The maximum protease activity was 64.12 units/ml, while the maximum bacterial count was 1.9 × 107 CFU/ml. The same was observed with B. licheniformis where the optimum bacterial growth was 1.7 × 108 CFU/ml and protease activity 82 units/ml to 37°C then decreased at 45°C. On the contrary, the growth of B. cereus and its activity gradually increased up to 45°C where reached to 5 × 107 CFU/ml and 116 units/ml.

- Monitoring protease activity (Units/ml) throughout the utilization process of casein soyabean broth using Bacillus subtilis,Bacillus licheniformis, and Bacillus cereus at 30, 37, and 45°C for 24 h.

- Monitoring the total bacterial count (log CFU/ml) throughout the utilization process of casein soyabean broth using Bacillus subtilis,Bacillus licheniformis, and Bacillus cereus at 30, 37, and 45°C for 24 h.
It was noticed that the enzyme activity and bacterial growth of B. subtilis and B. cereus strains were increased at alkaline medium. The maximum growth was 2 × 107 and 7.5 × 107 CFU/ml for both strains, respectively. However, the maximum activity was 67.5 and 56 units/ml, respectively. However, B. licheniformis gave the highest growth and activity at neutral pH with values 7 × 107 CFU/ml and 33.9 units/ml as demonstrated at [Figures 7a and b].

- Monitoring protease activity (Units/ml) throughout the utilization process of casein soyabean broth using Bacillus subtilis,Bacillus licheniformis, and Bacillus cereus at pH 4, 7, and 9 for 24 h at 30°C.
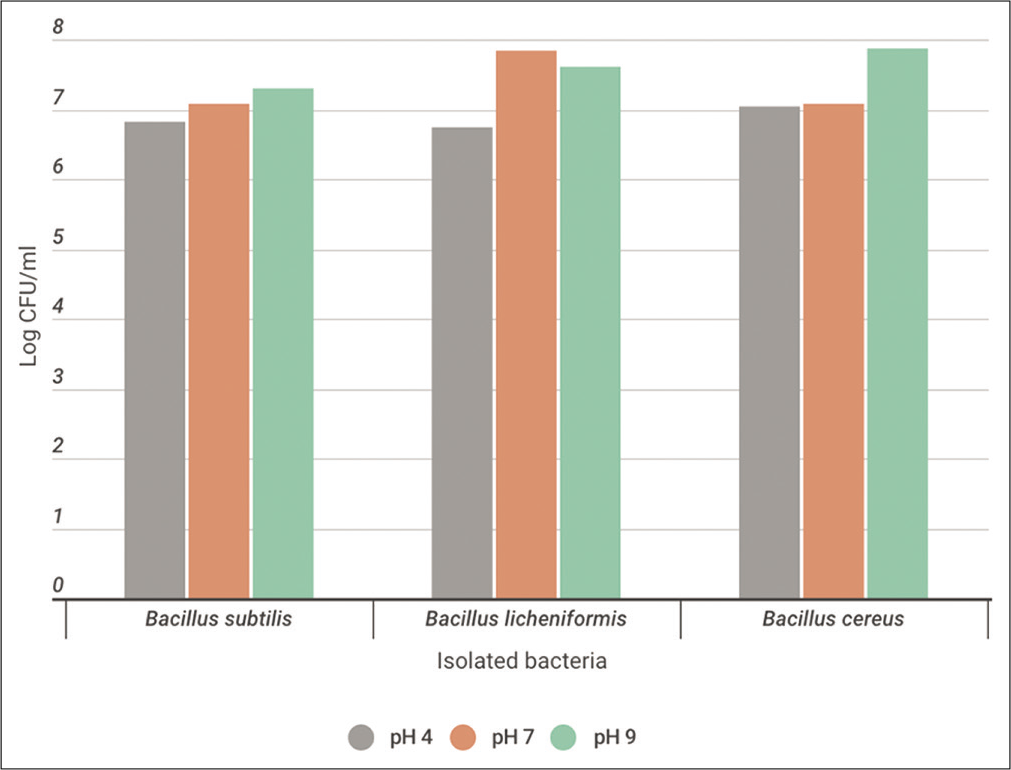
- Monitoring the total bacterial count (log CFU/ml) throughout the utilization process of casein soyabean broth using Bacillus subtilis,Bacillus licheniformis, and Bacillus cereus at pH 4, 7, and 9 for 24 h at 30°C.
In a trial to study the effect of salinity on the enzyme activity, it was found that the bacterial growth and enzyme activity of B. subtilis were reached to the maximum at 5% NaCl with values 2 × 107 CFU/ml and 67 units/ml, respectively. However, the maximum bacterial growth for B. licheniformis and B. cereus was at 2% NaCl. The bacterial count was 3.1 × 107 and 4.6 × 107 CFU/ml. The enzyme activity was maximum for B. lichinoformis at control culture with 33.9 units/ml and the maximum enzyme activity for B. cereus was at 2% NaCl with 106 units/ml. It was noticed that the growth of all strains was inhibited at 10% NaCl [Figures 8a and b].
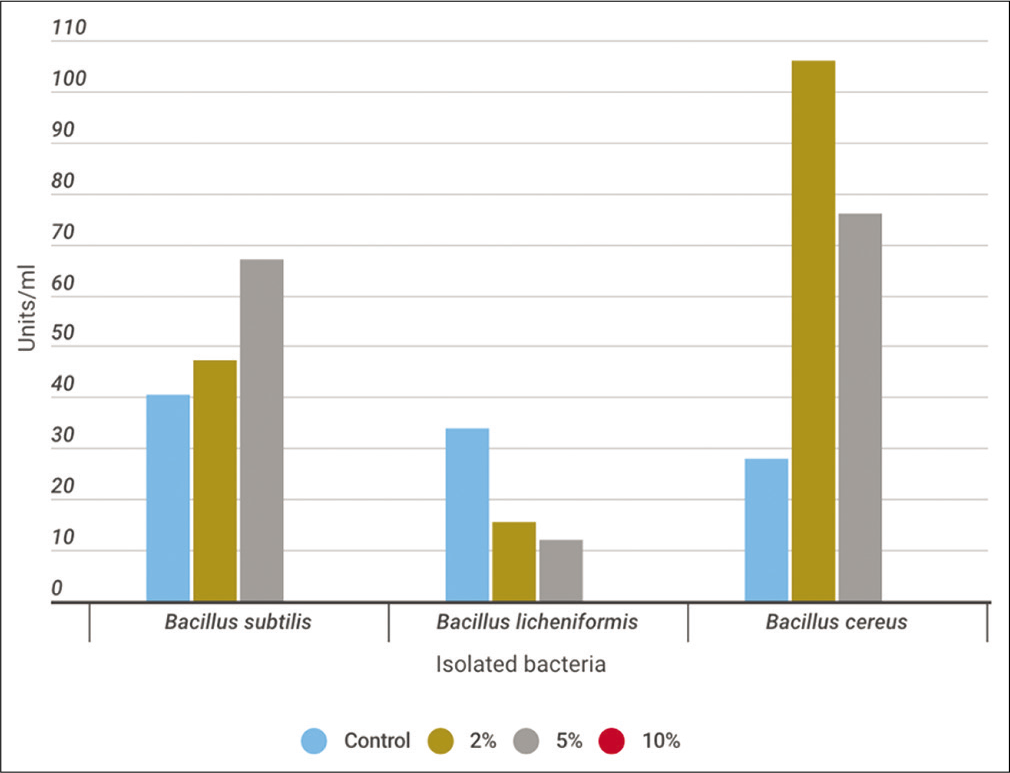
- Monitoring protease activity (Units/ml) throughout the utilization process of casein soyabean broth supplemented with 2, 5, and 10% NaCl using Bacillus subtilis,Bacillus licheniformis, and Bacillus cereus for 24 h at 30°C.
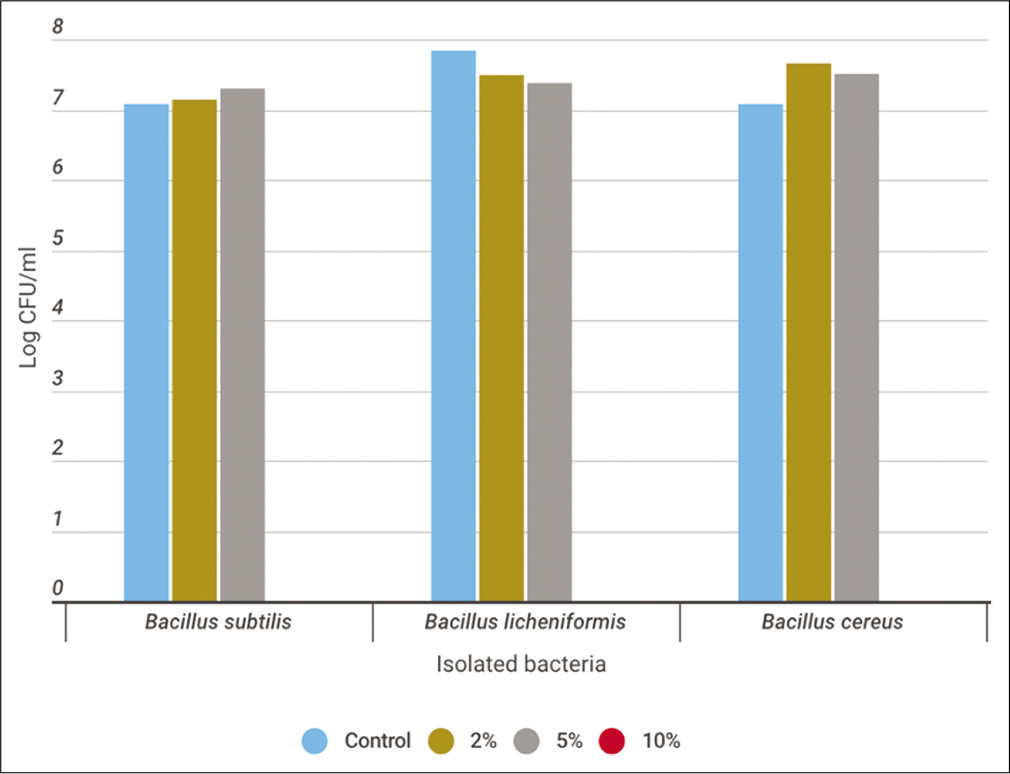
- Monitoring the total bacterial count (log CFU/ml) throughout the utilization process of casein soyabean broth supplemented with 2, 5, and 10% NaCl using Bacillus subtilis,Bacillus licheniformis, and Bacillus cereus for 24 h at 30°C.
Use of the extracted crude protease as antimicrobial agent
The sensitivity of three pathogenic bacteria against the extracellular crude protease enzyme produced from the three isolated bacteria was done. These pathogenic isolates were Gram-positive bacterial strain, and S. aureus. In addition, Gram-negative bacterial strains, E. coli, and K. pneumoniae.
In an attempt trial to test the susceptibility of the pathogenic bacteria against antibiotics, it was found that E. coli recorded resistance (40%) to the following antibiotics: AX, TM, E, CD, PG, VA, T, RP, Colistin, Fosfomycin, CTX, and AK, in addition, showed high sensitivity against the rest of antibiotics . S. aureus pathogen recorded high resistance (63%) to the following antibiotics: AX, TM, Cephalexin, CZ, CXM, Ceftriaxone, CTX, CAZ, CPM, ATM, Gentamicin, Tazobactam, IMI, MEM, NI, Colistin, CIP, CTX, and AK, in addition, showed sensitivity to the rest of antibiotics. Furthermore, K. pneumoniae recorded resistance (43%) to the following antibiotics: AX, TM, CAZ, NI, E, CD, PG, VA, T, RP, Colistin, and Fosfomycin, moreover, showed sensitivity to the rest of the tested antibiotics [Table 4].
| No. | Generic name | Conc. | Escherichia coli | Staphylococcus aureus | Klebsiella pneumoniae | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Status | Halo-zone mm | Status | Halo-zone mm | Status | Halo-zone mm | |||||
| 1 | Amoxicillin | 10 | R | 0 | R | 0 | R | 0 | ||
| 2 | Trimethoprim | 25 | R | 0 | R | 0 | R | 0 | ||
| 3 | Amoxicillin/Clavulanate | 30 | S | 26 | S | 28 | S | 28 | ||
| 4 | Cephalexin | 30 | S | 25 | R | 0 | S | 28 | ||
| 5 | Cefazolin | 30 | S | 25 | R | 0 | S | 28 | ||
| 6 | Cefuroxime | 30 | S | 25 | R | 0 | S | 28 | ||
| 7 | Ceftriaxone | 30 | S | 26 | R | 0 | S | 26 | ||
| 8 | Cefotaxime | 30 | S | 26 | R | 0 | S | 26 | ||
| 9 | Ceftazidime | 30 | S | 26 | R | 0 | R | -- | ||
| 10 | Cefepime | 30 | S | 26 | R | 0 | S | 28 | ||
| 11 | Aztreonam | 30 | S | 26 | R | 0 | S | 26 | ||
| 12 | Gentamicin | 10 | S | 22 | R | 0 | S | 24 | ||
| 13 | Amoxicillin | 10 | S | 24 | S | 35 | R | 0 | ||
| 14 | Tazobactam | 110 | S | 28 | R | 0 | S | 28 | ||
| 15 | Imipenem | 10 | S | 28 | R | 0 | S | 28 | ||
| 16 | Meropenem | 10 | S | 28 | R | 0 | S | 28 | ||
| 17 | Nitrofurantoin | 300 | S | 24 | R | 0 | R | -- | ||
| 18 | Colistin | 10 | S | 14 | R | 0 | S | 26 | ||
| 19 | Cefoxitin | 30 | S | 25 | S | 26 | S | 26 | ||
| 20 | Ciprofloxacin | 5 | S | 26 | R | 0 | S | 28 | ||
| 21 | Erythromycin | 15 | R | 0 | S | 26 | R | 0 | ||
| 22 | Clindamycin | 2 | R | 0 | S | 25 | R | 0 | ||
| 23 | Penicillin | 10 | R | 0 | S | 35 | R | 0 | ||
| 24 | Vancomycin | 30 | R | 0 | S | 20 | R | 0 | ||
| 25 | Tetracycline | 30 | R | 0 | S | 28 | R | 0 | ||
| 26 | Rifampicin | 5 | R | 0 | S | 28 | R | 0 | ||
| 27 | Cotrimoxazole/Colistin | 25 | R | 0 | S | 28 | R | 0 | ||
| 28 | Fosfomycin | 200 | R | 0 | S | 27 | R | 0 | ||
| 29 | Cefotaxime | 30 | R | 0 | R | 0 | S | 26 | ||
| 30 | Amikacin | 30 | R | 0 | R | 0 | S | 26 | ||
R: Resistance, S: Sensitive
As mentioned above, the activated isolated bacterial cells were inoculated to fresh Casein Soyabean Digest Broth. The new culture was allowed to grow overnight at 37°C with shaking. At the indicated time, CFU/ml was determined. In addition, 1.5 ml of the growing culture was taken. The supernatants were then used as crude enzyme to determine the protease activity, protein content as well as to test as antimicrobial agents. [Table 5] shows the bacterial growth (CFU/ml), protease activity (Units/ml), soluble protein content (μg/ml), and specific activity (Units/ml/mg/ml) for B. subtilis, B. licheniformis, and B. cereus which their extract were used as antimicrobial agents. [Table 5] shows that 1 μl crude enzyme extracted from B. subtilis, B. licheniformis, and B. cereus contain 0.073, 0.076, and 0.092 specific activity (Units/μl/μg/ml).
| Bacterial strain | Bacteria count | Protease activity | Soluble protein content (mg/ml) 1 | Specific activity |
|---|---|---|---|---|
| CFU/ml | Units/ml | (Units/ml/mg/ml) | ||
| B. subtilis | 2.7×107 | 40.2 | 0.546 | 73 |
| B. licheniformis | 3.2×107 | 46.6 | 0.608 | 76 |
| B. cereus | 6.8×106 | 49.7 | 0.54 | 92 |
1-Specific activity is dividing Enzyme activity (Units/ml)/Soluble protein (μg/ml). B. cereus: Bacillus cereus, B. licheniformis: Bacillus licheniformis, B. subtilis: Bacillus subtilis
Ten microliters of activated tested pathogen cells were inoculated to 10 ml of Casein Soyabean Digest Broth, supplemented with 0, 5, or 10 μl crude enzyme extract. All cultures were incubated O/N at 37°C. At the indicated time, measurement of O.D. of the turbidity at 600 nm was determined to detect the antimicrobial activity of the tested crude protease enzyme on the growth of pathogenic bacteria. [Figures 9-11] show a high effect on inhibiting the growth of pathogenic bacteria using 5 μl of crude enzyme with specific enzyme activity 73, 76, and 92 (Units/ml/mg/ml) for B. subtilis, B. licheniformis, and B. cereus, respectively. All pathogenic bacteria were totally inhibited with 10 μl more than 5 μl of crude enzyme.
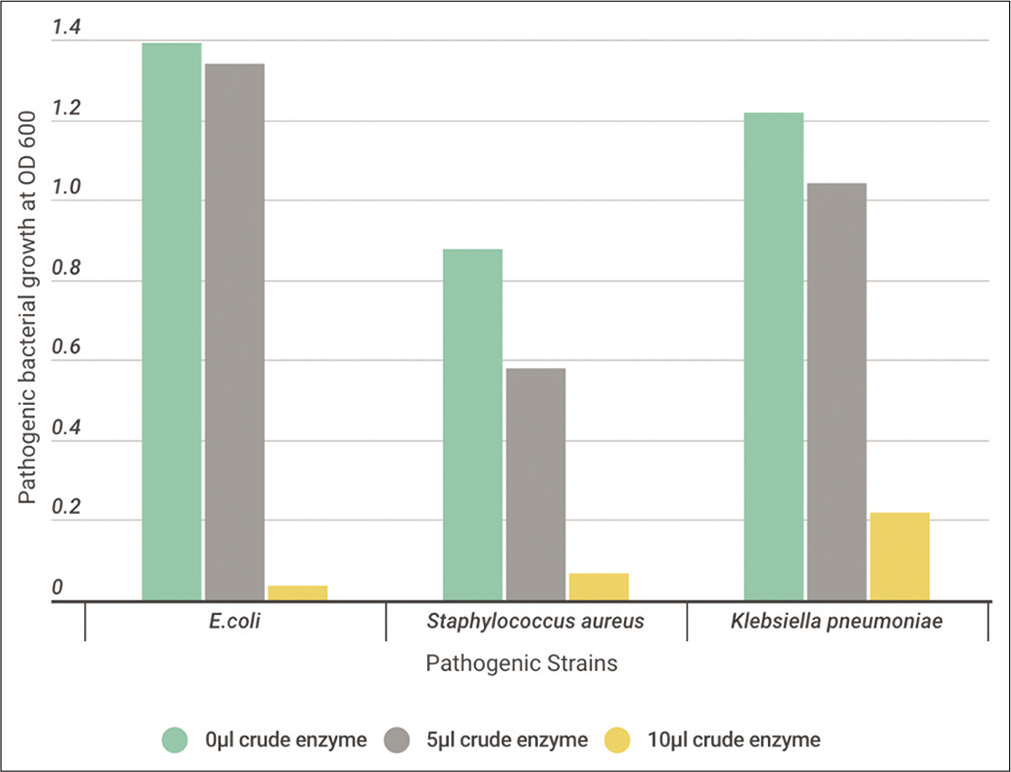
- The effect of different volumes (0, 5, and 10 μl) of crude protease enzyme extracted from Bacillus subtilis on the pathogenic strains.
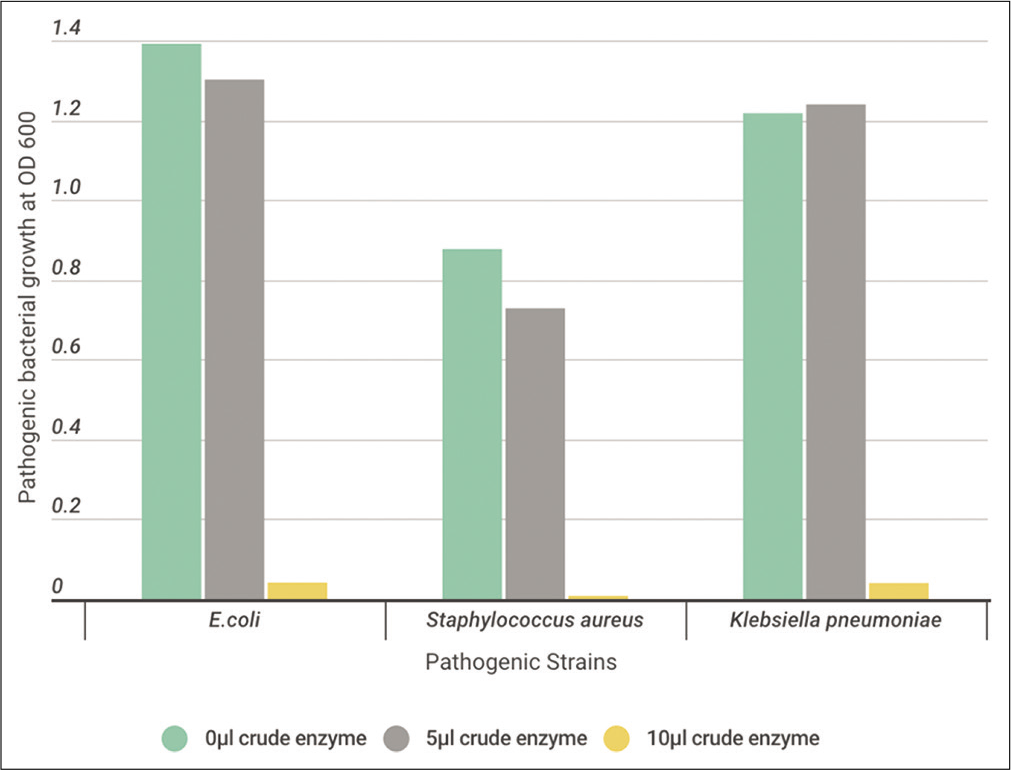
- The effect of different volumes (0, 5, and 10 μl) of crude protease enzyme extracted from Bacillus licheniformis on the pathogenic strains.
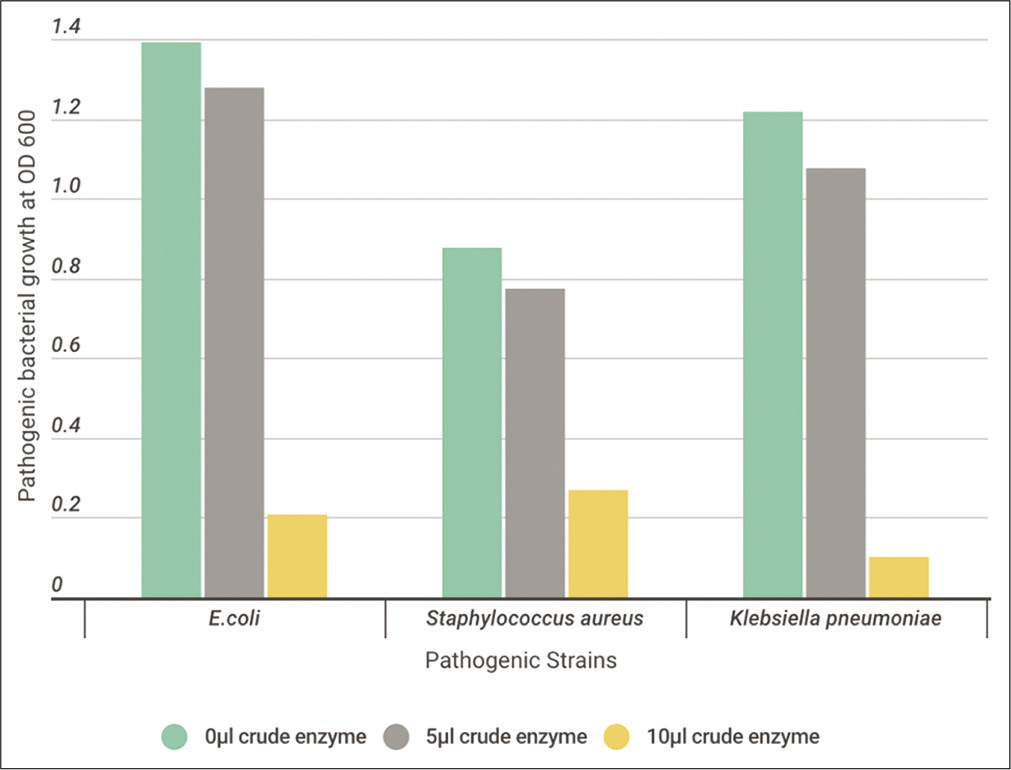
- The effect of different volumes (0, 5, and 10 μl) of crude protease enzyme extracted from Bacillus cereus on the pathogenic strains.
DISCUSSION
Bacteria are ubiquitous and highly diverse prokaryotes that can survive in adverse habitats. Thermophilic bacteria are microbes that mostly inhabit hot springs, live, and survive in temperatures between 45°C and 122°C. Thermophiles including bacterial and archaeal species are found in various geothermally heated regions of the earth such as hot springs and deep sea hydrothermal vents.[22] In the Kingdom of Saudi Arabia, there are ten geothermal springs with different deep temperatures of and different flow rates. They are distributed in Al-Lith and Gizan areas. Of these, Ain Khulab Quwa, Ain Khulab, Ain ad Damad, Ain Mijara Quwa, Ain al Wagrah, and Ain al Wagrah Dam are located in Gizan meanwhile Ain Jumah, Ain al Harra, Ain ad Darakah, and Ain Markub are located in Al-Lith area.[23]
Microbial produced enzymes that are secreted into the media are highly reliable for industrial processes and applications.[24] Protease enzymes are at the forefront of commercial enzymes as they have an industrial importance that makes up a large percentage of other enzymes. Commercially, it is used in various industrial food fields as softening of meat, bread, cheese, and beer and textiles industry as well as use in the field of pharmaceuticals, medical, and cosmetics production.[25]
In the current study, on the basis of maximum production of protease and gelatinase, three bacterial isolates were identified using 16S rRNA gene sequence analysis as B. cereus, and B. subtilis isolated from halophilic habitat, while B. licheniformis was isolated from thermophilic habitat (Al-Harth Hot spring). The selection of strains was depended on the choice Gram-positive and catalase-positive isolates. Therefore, they are strict aerobes or facultative anaerobes. The importance of Bacillus species in the industries was illustrated. Where, the genus Bacillus is probably the most important bacterial source of extracellular enzymes and is capable of producing high yields of alkaline and neutral proteolytic enzymes with remarkable properties, such as high stability toward extreme temperatures, pH, organic solvents, detergents, and oxidizing compounds.[26] In addition, Bacillus sp. can form endospore allowing it to tolerate harsh environmental conditions. It can secrete proteins into the medium, which is considered as a major advantage. This allows the accumulation of the native product with high yield in a pure form. Moreover, they have no known pathogenic interaction with animals or human.[27]
B. subtilis is a Gram-positive and catalase-positive bacteria found in the soil. Bacillus microflora has the ability to form a solid, protective microbial that allows it to withstand harsh environmental conditions.[28] In addition, B. licheniformis is a Gram-positive bacterium, often found in soil. These bacteria have the ability to live at high temperatures and are considered one of the most important types of bacteria producing industrial enzymes and that for their ability to produce alkali serine proteases, examples of industrial products are detergents.[29] Meanwhile, B. cereus is an endemic bacterium that lives in the soil. This is a human-causing strain that is transmitted through contaminated food. Harmless strains of wax Bacilli are used as feed additives in bio-nutrition to reduce salmonella in the intestines of animals. This improves the growth of animals and makes their meat safe and suitable for human consumption.[30]
Biochemical test reactions that are not universally positive or negative within a species may define biotypes of the species. Since bacteria will react differently to these tests, it is much like the bacterial “fingerprint.” To characterize the selected strains, the biochemical tests were done for all strains. They appeared utilization of some carbohydrates and other substrates as shown in the results. These results were compatible with the previous studies.[1,31]
The differences among strains may also be detected by variations in sensitivity to fixed concentrations of chemicals such as the antibiotic susceptibility test, where the antibiotics resistance in bacteria is a major health problem in many countries.[32] Resistance to antibiotics is acquired by a change in the gene makeup of bacterium, which can occur either by a gene mutation or by transfer of antibiotic resistance genes between bacteria in the environment.[33] The susceptibility of Bacilli to different antibiotics has been studied, and it has been demonstrated that in principle it should be possible to identify species based on the results of susceptibility tests. A large number of Bacillus strains assigned to different species were tested to determine their susceptibilities to antibiotics. Some clear differences between species were observed.[34] The antibiotic susceptibility test is performed to detect possible drug resistance and to assure susceptibility to drugs of choice for particular contamination.[1]
In the current study, bacterial isolates recorded high sensitivity with different degrees against tested antibiotics. In the previous studies, B. subtilis isolated from Al-khoba hot spring, Jazan, another B. subtilis isolated from tap water and B. thuringiensis were sensitive against most of tested antibiotics.[1,22,35] In a similar study, as part of a clinical-microbiological study, 89 strains of Bacillus spp. isolated from clinical blood cultures were tested for antimicrobial agent susceptibility to 18 antibiotics. MIC susceptibility tests revealed all B. cereus strains to be susceptible to IMI, VA, chloramphenicol, gentamicin, and CIP. Disk susceptibility testing suggested that B. cereus was rarely susceptible to PGs, semisynthetic PGs, or cephalosporins with the exception of mezlocillin.[36] In addition, the B. cereus isolates from cereals were susceptible to most of the antibiotics tested, but they were highly resistant to ampicillin, CPM, oxacillin, and PG.[37] In addition, the MIC for eight antibiotics was determined for 85 Bacillus species strains, B. subtilis subsp. subtilis (n = 29), B. licheniformis (n = 38), and B. sonorensis (n = 18). The antibiotic susceptibilities and characteristics of 94 B. licheniformis strains isolated from traditional Korean fermented soybean foods were assessed. The minimum inhibitory concentration (MIC) tests revealed that all strains were susceptible to gentamicin, kanamycin, T, and VA and that antibiotic resistances were expressed in a strain-specific manner. The resistances of B. licheniformis to chloramphenicol and streptomycin were established as intrinsic characteristics.[38]
In the current study, due to the importance of proteases in the industries, the extracellular protease production from B. cereus, B. licheniformis, and B. subtilis was chosen for further work. Due to the ever-increasing economic relevance of proteases, optimization of some fermentation parameters for Bacillus sp. was done. The effect of temperature, pH, and salinity on the bacterial growth and enzyme production was studied. Proteases production by microorganisms is affected by the medium compositions in addition to the environmental conditions. Thus, it is important to explore the influencing factors to attain the maximum protease production.[1] Cultivation temperature affects protein synthesis by influencing rate of biochemical reactions within the cell and consequently inducing or repressing enzyme production.[39]
The optimum temperature of the growth of B. subtilis and B. licheniformis was at 37°C. However, B. cereus showed high growth with protease activity at the optimum temperature of 45°C. In addition, it was found that the maximum growth with protease production for B. subtilis at 37°C.[40] Nevertheless, in another study of two isolates Bacilli that showed growth at high temperature 45°C.[41] In a previous study, B. licheniformis has been isolated and produced extracellular proteases and was able to grow at temperatures up to 60°C and at pH values up to 9.0 when casein was used as carbon source.[42]
pH affects the growth of microorganisms by particularly affecting the enzymes involved in the biosynthesis and growth process. For each microorganism, the growth rate is determined for a specific pH value. There is also a maximum value of maximum pH, which is the maximum value in which growth can be observed, and any increase in this value leads to preventing growth. Minimum pH is the lowest value in which growth can be observed.[43] Most natural environments have pH values between 5 and 9 and most organisms have pH optimal in this range. The protease enzyme showed activity in the wide range of pH from 6 to 11 with maximum protease activity at pH10.[44] In the current study, the protease enzyme activity and bacterial growth of B. subtilis and B. cereus strains were increased at alkaline medium. On the other hand, B. licheniformis gave the highest growth and activity at neutral pH. These results were agreeing with a previous study, where B. subtilis fermentation gave high level of protease production at 45°C after 36 h at pH-10.[45] In another study, the optimal pH and optimal temperature of enzyme production from Bacillus sp. strain were 8.0 and 45°C, respectively. Studies on enzymatic characterization revealed that crude protease showed maximum activity at pH 9.0 and 60°C, which is indicating the enzyme to be thermoalkaline protease.[2]
Proteases from halophilic microorganisms possess the advantage of being stable under high salinity levels. To maintain osmolality in saline environments, the microorganisms adopt mainly two strategies: One followed by most moderately halophilic bacteria, accumulating organic compatible solutes in the cytoplasm, and the second followed by the halobacteria, accumulating inorganic salts in the cytoplasm. In the current study, it showed that B. subtilis was halotolerant within concentrations at 5% NaCl, while B. licheniformis and B. cereus showed maximum growth at 2% NaCl. However, in a previous study, where the influence of NaCl concentrations on the fermentation process of Bacillus thuringensis isolated from mangrove rhizosphere to treat whey waste was investigated. Among the salinity supplemented media, the highest bacterial growth (log CFU/ ml 9.04) and maximum protease production (125.03 U/ml) were recorded in the presence of 10% NaCl.[1] In another study similar to the current study, Bacillus luteus H11 isolated from an alkaline soda lime was remarkably stable in the presence of Na Cl up to 5 M. The enzyme was active in a broad range of pH values and temperatures, with an optimum pH of 10.5 and a temperature of 45°C. The halo-alkaline protease produced by B. luteus H11 seems to be significant from an industrial perspective because of its tolerance towards high salinity and alkalinity as well as its stability against some organic solvents, surfactants, and oxidants.[46] Meanwhile, Protease producing halotolerant bacterium was isolated from saltern pond sediment and identified as B. licheniformis. The maximum protease production (141.46 U/mg) was obtained after addition of 1M NaCl.[47]
Antibiotic resistance appears because of evolution by natural selection. The effect of the antibiotic creates environmental pressure on the bacteria, but the mutations that appear in some bacterial cells cause them to escape the effect of the antibiotic. After that, this feature is transferred to the next offspring, which is characterized as a generation with full antibiotic resistance. Several studies have shown that the way antibiotics are used generally affects the evolution of the number of microorganisms resistant. The overuse of broad-spectrum antibiotics, such as second and third-generation cephalosporins, accelerates the development of methylene resistance. Other factors include inaccurate medical diagnosis, doctor prescribing unnecessary medications, inappropriate use of antibiotics by the patient, as well as the use of antibiotics as additional materials to feed and grow livestock.[40] Hence, bacterial proteases are an extensive collection of enzymes that have vital roles in cell viability, stress response, and pathogenicity. Their perturbation clearly offers the potential for antimicrobial drug development, both as traditional antibiotics and anti-virulence drugs.[48]
In this study, the susceptibility of three pathogenic bacteria against the extracellular crude protease enzyme produced from the three isolated bacteria was done. These pathogenic isolates were S. aureus, E. coli, and K. pneumoniae. A high effect on inhibiting the growth of pathogenic bacteria was registered. These results suggest that protease enzyme, could represent a promising antimicrobial drug strategy. In a similar study, 29 soil-isolated Bacillus species were tested against to some bacteria in terms of their general inhibition effects. Isolates have been found to be effective against Gram-positive and Gram-negative bacteria, while their extensive inhibition effect is particularly effective against gram-positive bacteria. In comparison, B. cereus strain has an inhibitory effect on Gram-positive as well as Gram-negative bacteria. In addition, certain isolates are more effective against test bacteria compared to other antibiotics.[49] Meanwhile, extracellular protease was isolated and purified from Xylaria psidii KT30 and used as antibacterial agent against Gram-positive bacteria; B. subtilis and S. aureus.[50] In another study, the effect of Bacillus pumilus proteolytic enzymes on the structure of 7-day-old S. marcescens biofilms was examined. A high efficacy of subtilisin-like protease and glutamyl endopeptidase in biofilm removal was noticed.[51] Furthermore, it was revealed the capacity of B. cereus to generate antimicrobial peptides from casein, through the production of extracellular enzymes, presents a new model of antagonistic competition.[52] B. subtilis VCDA that had proven antibacterial activity was screened for its production of bioactive extracellular proteases. In another study, the antibiofilm activity of the B. licheniformis protease enzymes was evaluated toward four pathogenic bacterial strains, S. aureus, Pseudomonas aeruginosa, E. coli, and B. subtilis, and the results showed that proteases from B. licheniformis might be useful for controlling biofilm formation by some pathogenic bacteria.[53]
CONCLUSION
In general, the extracellular crude protease enzyme of the isolated bacteria can able to use as antimicrobial agents against pathogenic bacterial strains. Further studies are recommended in future prospective. The recommended studies are the study of further factors affecting extracellular protease enzyme activity, scaling up of enzyme production, enzyme purification, studying the kinetics of enzyme, the effect of inhibitors, and immobilization of enzyme. Focusing on the application as biology control on a large number of pathogenic bacteria and fungi with different concentrations to determine MICs with a comparison with the effect of antibiotics is recommended.
Declaration of patient consent
Patient’s consent not required, as there are no patients in this study.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Whey fermentation for protease production using Bacillus thuringiensis isolated from mangrove rhizosphere soil in Jazan, Saudi Arabia. Pol J Environ Stud. 2020;29:1-10.
- [CrossRef] [Google Scholar]
- Effect of culture conditions on the production of an extracellular protease by Bacillus sp isolated from Soil Sample of Lavizan Jungle Park. Enzyme Res. 2011;2011:219628.
- [CrossRef] [PubMed] [Google Scholar]
- Antibacterial effect of extracts of Ocimum gratissimum and Piper guineense on Escherichia coli and Staphylococcus aureus. Afr J Food Sci. 2009;3:77-81.
- [Google Scholar]
- Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous hexosaminidase activity. J Bacteriol. 2003;185:4693-8.
- [CrossRef] [Google Scholar]
- Microbial proteases in commercial applications. J Pharm Chem Biol Sci. 2016;4:365-74.
- [Google Scholar]
- Laboratory Exercises in Microbiology (4th ed). New York: McGraw Hill. Inc.; 1977. p. :200-60.
- [Google Scholar]
- Recycling of keratin containing materials (chicken feather) through genetically engineered bacteria. Polym Plast Technol Eng. 2004;43:1589-99.
- [CrossRef] [Google Scholar]
- Gelatin Hydrolysis Test: Principle, Procedure and Expected Results, Medical Microbiology Guide, Microbeonline 2014.
- [Google Scholar]
- Catalase Test: Principle, Procedure, Results and Applications, Biochemical tests in Microbiology, Microbiology for Beginners, Medical Microbiology Guide, Microbeonline 2013.
- [Google Scholar]
- Identification System for Streptococcaceae and Related Organisms. API 20 Strep Analytical Profile;.
- [Google Scholar]
- Bacteriological analysis, antimicrobial susceptibility and detection of 16S rRNA. Malays J Microbiol. 2009;5:123-27.
- [Google Scholar]
- Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990;28:1942-6.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular Cloning: A Laboratory Manual (3rd ed). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2001. p. :2100.
- [Google Scholar]
- Purification of alkaline protease from hydrolyzed chicken feather waste using recombinant B. subtilis strain. Sci J King Faisal Univ. 2012;13:1433.
- [Google Scholar]
- Production of alkaline protease by immobilized mycelium of Conidiobolus. Enzyme Microb Technol. 1986;8:632-4.
- [CrossRef] [Google Scholar]
- Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76-85.
- [CrossRef] [Google Scholar]
- Antibacterial activity of extracellular protease isolated from an Algicolous fungus Xylaria psidii KT30 against gram-positive bacteria. HAYATI J Biosci. 2016;23:73-8.
- [CrossRef] [Google Scholar]
- Production of protease showing antibacterial activity by Bacillus subtilis VCDA associated with tropical marine sponge Callyspongia diffusa. J Microb Biochem Technol. 2017;10:1948-5948.
- [Google Scholar]
- Characterization of bacteria isolated from domestic wastewater in Jazan, Saudi Arabia. Egypt J Exp Biol Bot. 2018;14:331-8.
- [Google Scholar]
- Characterization of thermophilic bacteria isolated from two hot springs in Saudi Arabia. J Pure Appl Microbiol. 2017;11:743-52.
- [CrossRef] [Google Scholar]
- Geothermal Resources of Saudi Arabia-Country Update Report. Proceedings World Geothermal Congress.
- Biotechnological processes in microbial amylase production. Biomed Res Int. 2017;2017:1272193.
- [CrossRef] [PubMed] [Google Scholar]
- Application of protease technology in dermatology. J Clin Aesthet Dermatol. 2013;6:14-22.
- [Google Scholar]
- An overview of Bacillus proteases: From production to application. Crit Rev Biotechnol. 2018;38:321-34.
- [CrossRef] [PubMed] [Google Scholar]
- Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117-26.
- [CrossRef] [PubMed] [Google Scholar]
- Memory and modularity in cell-fate decision Maki. Nature. 2013;503:481-6.
- [CrossRef] [PubMed] [Google Scholar]
- Nitrogen catabolite repression of the L-asparaginase of Bacillus licheniformis. J Bacteriol. 1985;164:938-40.
- [CrossRef] [PubMed] [Google Scholar]
- Bacillus cereus, a volatile human pathogen. Clin Microbiol Rev. 2010;23:382-98.
- [CrossRef] [PubMed] [Google Scholar]
- A review on commonly used biochemical test for bacteria. Innov J Life Sci. 2013;1:1-7.
- [Google Scholar]
- PCR targeting of antibiotic resistant bacteria in public drinking water of Lahore metropolitan, Pakistan. Biomed Environ Sci. 2009;22:458-63.
- [CrossRef] [Google Scholar]
- Studies on antibiotics and heavy metal resistance profiling of Escherichia coli from drinking water and clinical specimens. Biosci Disscov. 2012;3:292-5.
- [Google Scholar]
- Antibiotic susceptibility testing of isolated Bacillus thuringiensis from three soil types around Iligan City, Philippines. Afr J Microbiol Res. 2013;7:678-82.
- [Google Scholar]
- Isolation, identification and characterization of Bacillus subtilis from tap water. Asian J Microbiol Biotechnol Environ Sci. 2017;19:817-30.
- [Google Scholar]
- In vitro susceptibility of Bacillus spp. to selected antimicrobial agents. Antimicrob Agents Chemother. 1988;32:642-5.
- [CrossRef] [PubMed] [Google Scholar]
- Enterotoxin genes, antibiotic susceptibility, and biofilm formation of low-temperature-tolerant Bacillus cereus isolated from green leaf lettuce in the cold chain. Foods. 2009;9:249.
- [CrossRef] [PubMed] [Google Scholar]
- Antibiotic susceptibilities and characteristics of Bacillus licheniformis isolates from traditional Korean fermented soybean foods. LWT. 2017;75:565-8.
- [CrossRef] [Google Scholar]
- Relationship of critical temperature to macromolecular synthesis and growth yield in Psychrobacter cryopegella. J Bacteriol. 2004;186:2340-5.
- [CrossRef] [PubMed] [Google Scholar]
- Study of protease enzyme from Bacillus species and its application as a contact lens cleanser. Br Biomed Bull. 2014;2:292-302.
- [Google Scholar]
- Thermostable alkaline proteases of Bacillus licheniformis MIR 29: Isolation, production and characterization. Appl Microbiol Biotechnol. 1994;45:327-32.
- [CrossRef] [Google Scholar]
- Optimizing Some Factors Affecting Alkaline Protease Production by a Marine Bacterium Streptomyces albidoflavus. Proceeding of 5th Scientific Environmental Conference. Zagazig University.
- Production and characterization of alkaline protease from a high yielding and moderately halophilic strain of SD11 marine bacteria. . 2015;2015:798304.
- [CrossRef] [Google Scholar]
- Production, optimization and partial purification of protease from Bacillus subtilis. J Taibah Univ Sci. 2014;9:150-60.
- [Google Scholar]
- Alkaline and halophilic protease production by Bacillus luteus H11 and its potential industrial applications. Food Technol Biotechnol. 2018;56:553-61.
- [CrossRef] [PubMed] [Google Scholar]
- Screening and optimization of protease production from a halotolerant Bacillus licheniformis isolated from saltern sediments. J Gen Eng Biotechnol. 2013;11:47-52.
- [CrossRef] [Google Scholar]
- Bacterial proteases, untapped antimicrobial drug targets. J Antibiot. 2017;70:366-77.
- [CrossRef] [PubMed] [Google Scholar]
- Antimicrobial activities of some Bacillus spp. strains isolated from the soil. Microbiol Res. 2006;161:127-31.
- [CrossRef] [PubMed] [Google Scholar]
- Antibacterial activity of extracellular protease isolated from an Algicolous Fungus Xylaria psidii KT30 against gram-positive bacteria. HAYATI J Biosci. 2016;23:73-8.
- [CrossRef] [Google Scholar]
- Effects of Bacillus serine proteases on the bacterial biofilms. Biomed Res Int. 2017;2017:8525912.
- [CrossRef] [PubMed] [Google Scholar]
- Two new secreted proteases generate a casein-derived antimicrobial peptide in Bacillus cereus food born isolate leading to bacterial competition in milk. Front Microbiol. 2018;9:1148.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characterization of alkaline protease-coding gene from Bacillus licheniformis MK90 mutants with biofilm inhibitory activity. Egypt Pharm J. 2019;18:419-33.
- [CrossRef] [Google Scholar]






